A comparison of single-cell trajectory inference methods: towards more accurate and robust tools
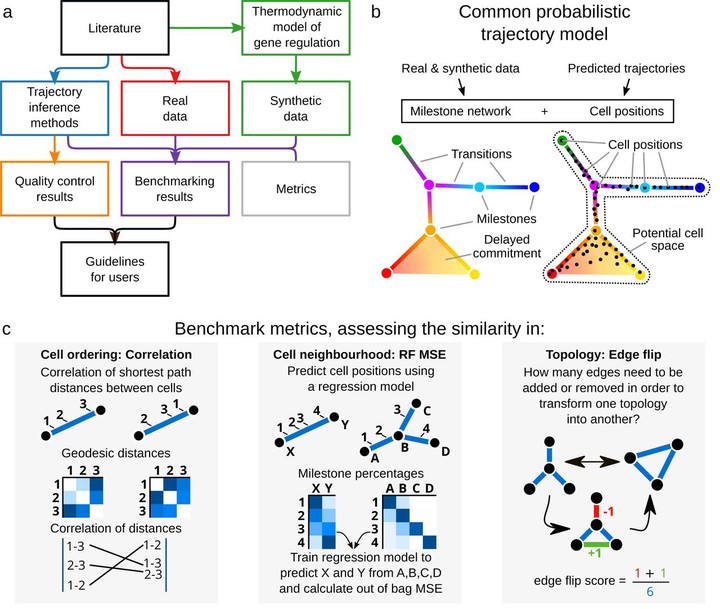 An overview of several key aspects of the evaluation.
An overview of several key aspects of the evaluation.Abstract
Using single-cell-omics data, it is now possible to computationally order cells along trajectories, allowing the unbiased study of cellular dynamic processes. Since 2014, more than 50 trajectory inference methods have been developed, each with its own set of methodological characteristics. As a result, choosing a method to infer trajectories is often challenging, since a comprehensive assessment of the performance and robustness of each method is still lacking. In order to facilitate the comparison of the results of these methods to each other and to a gold standard, we developed a global framework to benchmark trajectory inference tools. Using this framework, we compared the trajectories from a total of 29 trajectory inference methods, on a large collection of real and synthetic datasets. We evaluate methods using several metrics, including accuracy of the inferred ordering, correctness of the network topology, code quality and user friendliness. We found that some methods, including Slingshot, TSCAN and Monocle DDRTree, clearly outperform other methods, although their performance depended on the type of trajectory present in the data. Based on our benchmarking results, we therefore developed a set of guidelines for method users. However, our analysis also indicated that there is still a lot of room for improvement, especially for methods detecting complex trajectory topologies. Our evaluation pipeline can therefore be used to spearhead the development of new scalable and more accurate methods, and is available at github.com/dynverse/dynverse. To our knowledge, this is the first comprehensive assessment of trajectory inference methods. For now, we exclusively evaluated the methods on their default parameters, but plan to add a detailed parameter tuning procedure in the future. We gladly welcome any discussion and feedback on key decisions made as part of this study, including the metrics used in the benchmark, the quality control checklist, and the implementation of the method wrappers. These discussions can be held at github.com/dynverse/dynverse/issues.