 Showcase of dyngen functionality.
Showcase of dyngen functionality.Abstract
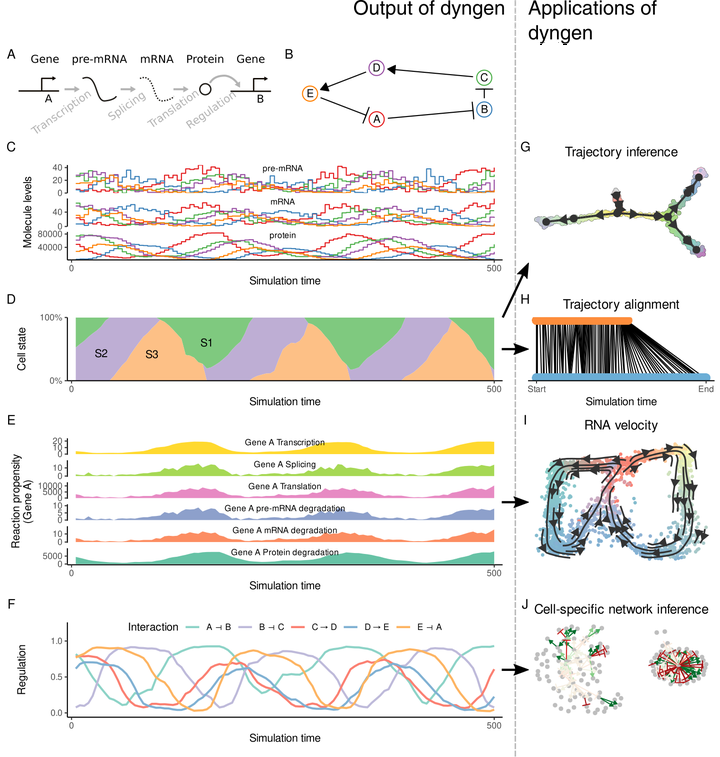
When developing new types of tools for single-cell analyses, there is often a lack of datasets on which to quantitatively assess the performance. We developed dyngen, a multi-modality simulator of single cells. In dyngen, the biomolecular state of an in silico changes over time according to a predefined gene regulatory network. We used dyngen to benchmark three emerging ways of analysing single-cell data: RNA velocity, cell-specific network inference and trajectory alignment methods. dyngen lays the foundations for benchmarking a wide variety of computational single-cell tools and can be used to help kick-start the development of future types of analyses.
Type
Publication
bioRxiv