SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development
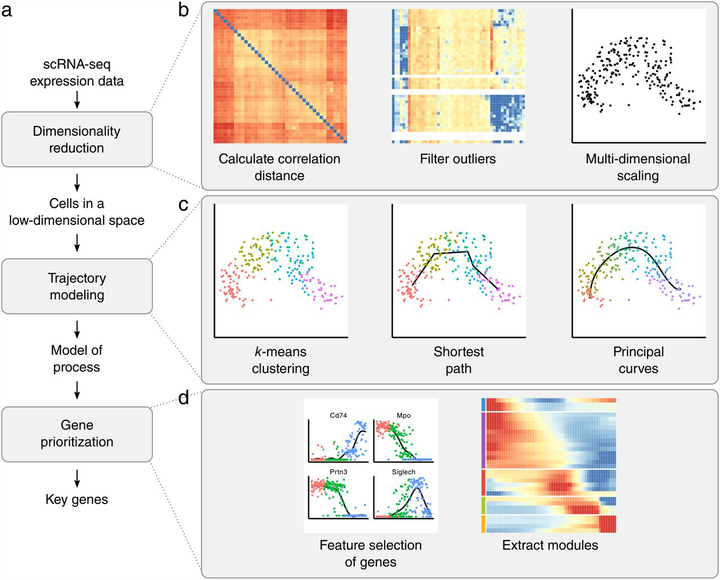 SCORPIUS infers a trajectory in three steps.
SCORPIUS infers a trajectory in three steps.Abstract
Recent advances in RNA sequencing enable the generation of genome-wide expression data at the single-cell level, opening up new avenues for transcriptomics and systems biology. A new application of single-cell whole-transcriptomics is the unbiased ordering of cells according to their progression along a dynamic process of interest. We introduce SCORPIUS, a method which can effectively reconstruct an ordering of individual cells without any prior information about the dynamic process. Comprehensive evaluation using ten scRNA-seq datasets shows that SCORPIUS consistently outperforms state-of-the-art techniques. We used SCORPIUS to generate novel hypotheses regarding dendritic cell development, which were subsequently validated in vivo. This work enables data-driven investigation and characterization of dynamic processes and lays the foundation for objective benchmarking of future trajectory inference methods.