The RNA Atlas, a single nucleotide resolution map of the human transcriptome
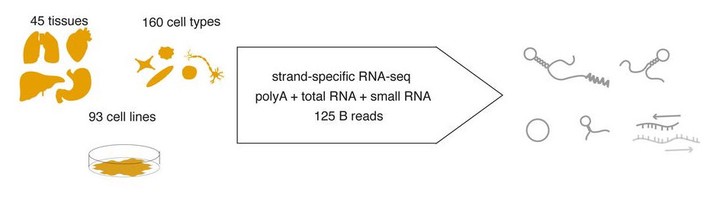 A heterogeneous collection of tissues, cell types and cell lines was sequenced through complementary strand-specific RNA-sequencing methods to profile the major RNA-biotypes in the human transcriptome.
A heterogeneous collection of tissues, cell types and cell lines was sequenced through complementary strand-specific RNA-sequencing methods to profile the major RNA-biotypes in the human transcriptome.Abstract
The human transcriptome consists of various RNA biotypes including multiple types of non-coding RNAs (ncRNAs). Current ncRNA compendia remain incomplete partially because they are almost exclusively derived from the interrogation of small- and polyadenylated RNAs. Here, we present a more comprehensive atlas of the human transcriptome that is derived from matching polyA-, total-, and small-RNA profiles of a heterogeneous collection of nearly 300 human tissues and cell lines. We report on thousands of novel RNA species across all major RNA biotypes, including a hitherto poorly-cataloged class of non-polyadenylated single-exon long non-coding RNAs. In addition, we exploit intron abundance estimates from total RNA-sequencing to test and verify functional regulation by novel non-coding RNAs. Our study represents a substantial expansion of the current catalogue of human ncRNAs and their regulatory interactions. All data, analyses, and results are available in the R2 web portal and serve as a basis to further explore RNA biology and function.