anndata for R has a new home!
Welcome to the dynverse project :)
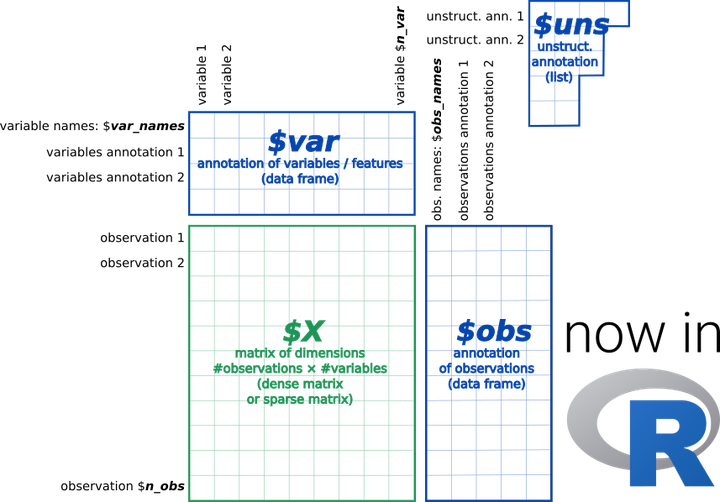 anndata provides a scalable way of keeping track of data and learned annotations.
anndata provides a scalable way of keeping track of data and learned annotations.anndata for R is brings h5ad processing to R with the same
easy-to-use interface as the Python anndata API.
No longer do you have to fiddle with hdf5r, reticulate or one of the
many conversion
functions.
The code base for anndata for R has been moved from rcannood/anndata to dynverse/anndata and with it got a fancy new homepage to be able to browse the documentation from: anndata.dynverse.org!
If you haven’t yet, please give anndata for R a try! We’ve found that by using anndata for R, interacting with other anndata-based Python packages becomes super easy¹! Below is a small demonstration.
Download and load dataset
Let’s use a 10x dataset from the 10x genomics website. You can download it to an anndata object with scanpy as follows:
library(anndata)
library(reticulate)
sc <- import("scanpy")
url <- "https://cf.10xgenomics.com/samples/cell-exp/6.0.0/SC3_v3_NextGem_DI_CellPlex_CSP_DTC_Sorted_30K_Squamous_Cell_Carcinoma/SC3_v3_NextGem_DI_CellPlex_CSP_DTC_Sorted_30K_Squamous_Cell_Carcinoma_count_sample_feature_bc_matrix.h5"
ad <- sc$read_10x_h5("dataset.h5", backup_url = url)
ad## AnnData object with n_obs × n_vars = 5377 × 36601
## var: 'gene_ids', 'feature_types', 'genome'Preprocessing dataset
The resuling dataset is a wrapper for the Python class but behaves very much like an R object:
ad[1:5, 3:5]## View of AnnData object with n_obs × n_vars = 5 × 3
## var: 'gene_ids', 'feature_types', 'genome'dim(ad)## [1] 5377 36601You can still call scanpy functions on it, for example to perform preprocessing.
sc$pp$filter_cells(ad, min_genes = 200)
sc$pp$filter_genes(ad, min_cells = 3)
sc$pp$normalize_per_cell(ad)
sc$pp$log1p(ad)Analysing your dataset in R
You can seamlessly switch back to using your dataset with other R functions. For example, calculating the rowMeans of the expression matrix.
library(Matrix)
rowMeans(ad$X[1:10,])## AAACCCAAGCGCGTTC-1 AAACCCAAGGCAATGC-1 AAACCCAGTATCTTCT-1 AAACCCAGTGACAACG-1
## 0.05451418 0.13627126 0.12637224 0.13958617
## AAACCCAGTTGAATCC-1 AAACCCATCGGCTTGG-1 AAACGAAAGAGAGCCT-1 AAACGAAAGCTTAAGA-1
## 0.05979424 0.11365747 0.05011727 0.14347849
## AAACGAAAGGCACGAT-1 AAACGAAAGGTAGCCA-1
## 0.12979302 0.12366312Additional thoughts
¹ When it works. While anndata for R has certainly been useful for us, there is still a lot left to implement. For example, using h5ad-backed AnnData objects does not work yet. If you do encounter an issue, let us know by means of a GitHub Issue. Make sure to include a reproducible example!